Note
This tutorial was generated from a Jupyter notebook that can be downloaded here.
Analyzing Open MDC data
In this tutorial we will use enterprise to analyze open MDC dataset
1.
Get par and tim files
The first step in the process is getting the open MDC1 par and tim files in the tests directory.
parfiles = sorted(glob.glob(datadir + '/*.par'))
timfiles = sorted(glob.glob(datadir + '/*.tim'))
Load pulsars into Pulsar objects
enterprise uses a specific
Pulsar object to
store all of the relevant pulsar information (i.e. TOAs, residuals,
error bars, flags, etc) from the timing package. Eventually
enterprise will support both PINT and tempo2; however, for
the moment it only supports tempo2 through the
libstempo package. This object
is then used to initalize Signals that define the generative model
for the pulsar residuals. This is in keeping with the overall
enterprise philosophy that the pulsar data should be as loosley
coupled as possible to the pulsar model.
psrs = []
for p, t in zip(parfiles, timfiles):
psr = Pulsar(p, t)
psrs.append(psr)
Setup and run a simple noise model on a single pulsar
Here we will demonstrate how to do a simple noise run on a single pulsar. In this analysis we will simply model the noise via a single EFAC parameter and a power-law red noise process.
Set up model
Here we see the basic enterprise model building steps:
Define parameters and priors (This makes use of the Parameter class factory)
Set up the signals making use of the
Signalclass factories.Define the model by summing the individual
Signalclasses.Define a PTA by initializing the signal model with a
Pulsarobject.
Notice that powerlaw is uses as a
Function
here.
##### parameters and priors #####
# Uniform prior on EFAC
efac = parameter.Uniform(0.1, 5.0)
# red noise parameters
# Uniform in log10 Amplitude and in spectral index
log10_A = parameter.Uniform(-18,-12)
gamma = parameter.Uniform(0,7)
##### Set up signals #####
# white noise
ef = white_signals.MeasurementNoise(efac=efac)
# red noise (powerlaw with 30 frequencies)
pl = utils.powerlaw(log10_A=log10_A, gamma=gamma)
rn = gp_signals.FourierBasisGP(spectrum=pl, components=30)
# timing model
tm = gp_signals.TimingModel()
# full model is sum of components
model = ef + rn + tm
# initialize PTA
pta = signal_base.PTA([model(psrs[0])])
We can see which parameters we are going to be searching over with:
print(pta.params)
["J0030+0451_efac":Uniform(0.1,5.0), "J0030+0451_gamma":Uniform(0,7), "J0030+0451_log10_A":Uniform(-18,-12)]
Get initial parameters
We will start our MCMC chain at a random point in parameter space. We
accomplish this by setting up a parameter dictionary using the name
and sample methods for each Parameter.
xs = {par.name: par.sample() for par in pta.params}
print(xs)
{u'J0030+0451_efac': 4.7352650698633516, u'J0030+0451_gamma': 3.8216965873513029, u'J0030+0451_log10_A': -15.161366939011094}
Note that the rest of the analysis here is dependent on the sampling
method and not on enterprise itself.
Set up sampler
Here we are making use of the
PTMCMCSampler package
for sampling. For this sampler, as in many others, it requires a
function to compute the log-likelihood and log-prior given a vector of
parameters. Here, these are supplied by PTA as
pta.get_lnlikelihood and pta.get_lnprior.
# dimension of parameter space
ndim = len(xs)
# initial jump covariance matrix
cov = np.diag(np.ones(ndim) * 0.01**2)
# set up jump groups by red noise groups
ndim = len(xs)
groups = [range(0, ndim)]
groups.extend([[1,2]])
# intialize sampler
sampler = ptmcmc(ndim, pta.get_lnlikelihood, pta.get_lnprior, cov, groups=groups,
outDir='chains/mdc/open1/')
Sample!
# sampler for N steps
N = 100000
x0 = np.hstack(p.sample() for p in pta.params)
sampler.sample(x0, N, SCAMweight=30, AMweight=15, DEweight=50)
Finished 10.00 percent in 7.578883 s Acceptance rate = 0.27876Adding DE jump with weight 50
Finished 99.00 percent in 77.849424 s Acceptance rate = 0.404505
Run Complete
Examine chain output
We see here that we have indeed recovered the injected values!
chain = np.loadtxt('chains/mdc/open1/chain_1.txt')
pars = sorted(xs.keys())
burn = int(0.25 * chain.shape[0])
truths = [1.0, 4.33, np.log10(5e-14)]
corner.corner(chain[burn:,:-4], 30, truths=truths, labels=pars);

Run full PTA GWB analysis
Here we will use the full 36 pulsar PTA to conduct a search for the GWB.
In this analysis we fix the EFAC=1 for simplicity (and since we already
know the answer!). This shows an example of how to use Constant
parameters in enterprise.
Here you notice some of the simplicity of enterprise. For the most
part, setting up the model for the full PTA is identical to that for one
pulsar. In this case the only differences are that we are specifying the
timespan to use when setting the GW and red noise frequencies and we are
including a FourierBasisCommonGP signal, which models the GWB
spectrum and spatial correlations.
After this setup, the rest is nearly identical to the single pulsar run above.
# find the maximum time span to set GW frequency sampling
tmin = [p.toas.min() for p in psrs]
tmax = [p.toas.max() for p in psrs]
Tspan = np.max(tmax) - np.min(tmin)
##### parameters and priors #####
# white noise parameters
# in this case we just set the value here since all efacs = 1
# for the MDC data
efac = parameter.Constant(1.0)
# red noise parameters
log10_A = parameter.Uniform(-18,-12)
gamma = parameter.Uniform(0,7)
##### Set up signals #####
# white noise
ef = white_signals.MeasurementNoise(efac=efac)
# red noise (powerlaw with 30 frequencies)
pl = utils.powerlaw(log10_A=log10_A, gamma=gamma)
rn = gp_signals.FourierBasisGP(spectrum=pl, components=30, Tspan=Tspan)
# gwb
# We pass this signal the power-law spectrum as well as the standard
# Hellings and Downs ORF
orf = utils.hd_orf()
crn = gp_signals.FourierBasisCommonGP(pl, orf, components=30, name='gw', Tspan=Tspan)
# timing model
tm = gp_signals.TimingModel()
# full model is sum of components
model = ef + rn + tm + crn
# initialize PTA
pta = signal_base.PTA([model(psr) for psr in psrs])
Set up sampler
# initial parameters
xs = {par.name: par.sample() for par in pta.params}
# dimension of parameter space
ndim = len(xs)
# initial jump covariance matrix
cov = np.diag(np.ones(ndim) * 0.01**2)
# set up jump groups by red noise groups
ndim = len(xs)
groups = [range(0, ndim)]
groups.extend(map(list, zip(range(0,ndim,2), range(1,ndim,2))))
sampler = ptmcmc(ndim, pta.get_lnlikelihood, pta.get_lnprior, cov, groups=groups,
outDir='chains/mdc/open1_gwb/')
# sampler for N steps
N = 100000
x0 = np.hstack(p.sample() for p in pta.params)
sampler.sample(x0, N, SCAMweight=30, AMweight=15, DEweight=50)
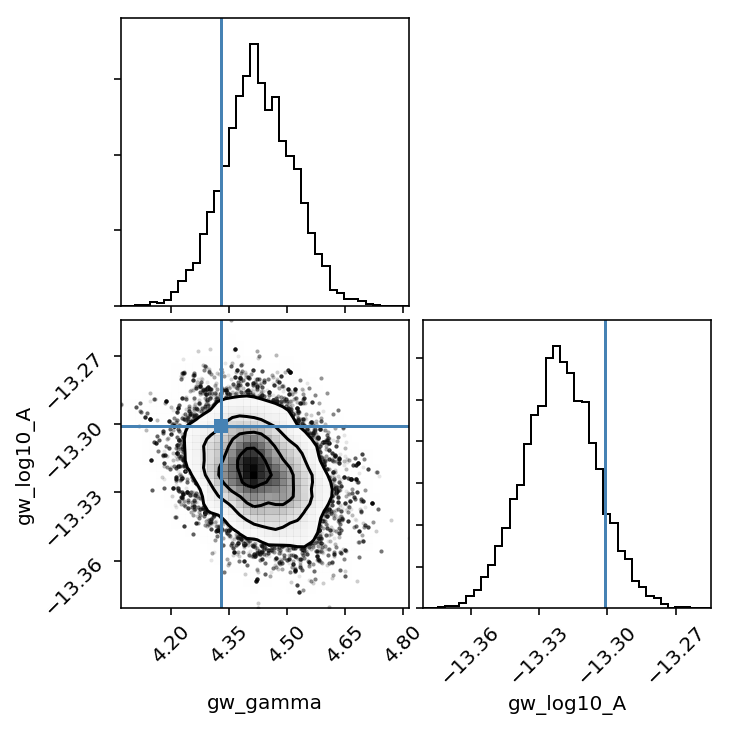
Plot output
chain = np.loadtxt('chains/mdc/open1_gwb/chain_1.txt')
pars = sorted(xs.keys())
burn = int(0.25 * chain.shape[0])
corner.corner(chain[burn:,-6:-4], 40, labels=pars[-2:], smooth=True, truths=[4.33, np.log10(5e-14)]);